Three-dimensional (3D) image segmentation is encountered in scientific studies of z-stacks acquired by confocal laser scanning microscopes. Our objectives are (a) to automate segmentation of a large number of 3D z-stacks and (b) to estimate the segmentation accuracy from projected ground truth, statistical samples and visual inspection inputs.
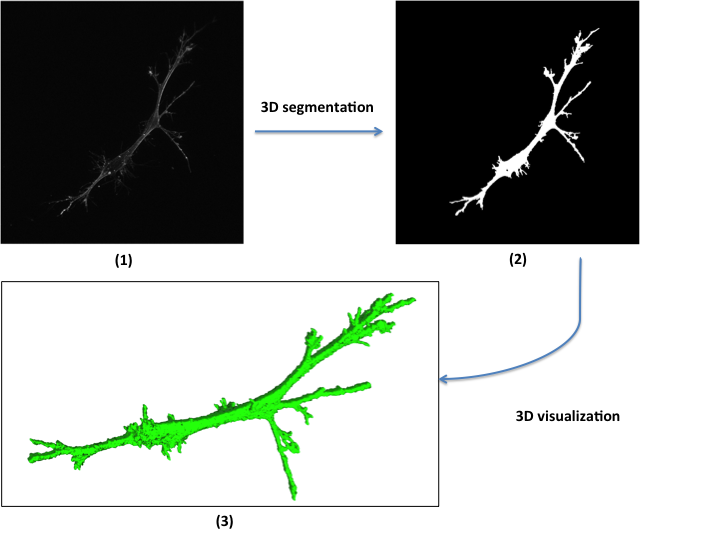
Human bone marrow stromal cells were cultured on 10 different scaffolds and imaged in 3D using confocal laser scanning microscopy. Approximately 110 cells were imaged per scaffold yielding 1100 Z-stacks, 122,949 images and 122 GB of data. Six segmentation processes were designed and the most accurate method was selected upon imaging and geometrical assumptions. Selection was driven by minimization of human labor needed to provide ground truth while maximizing statistical confidence in the segmentation accuracy.
Segmentation accuracy was determined against manually segmented z-stacks. The two most accurate segmentation methods were applied to all z-stacks and visually inspected in a 2D tiled view and interactive 3D view in a web browser. The results of visual inspections are incorporated into the analyses to increase the confidence in segmentation accuracy estimates.
We built a 3D web-based visualization tool allowing the visualization of 3D volumes representing the segmented z-stacks.
This tool is accessible online by clicking on the following link:
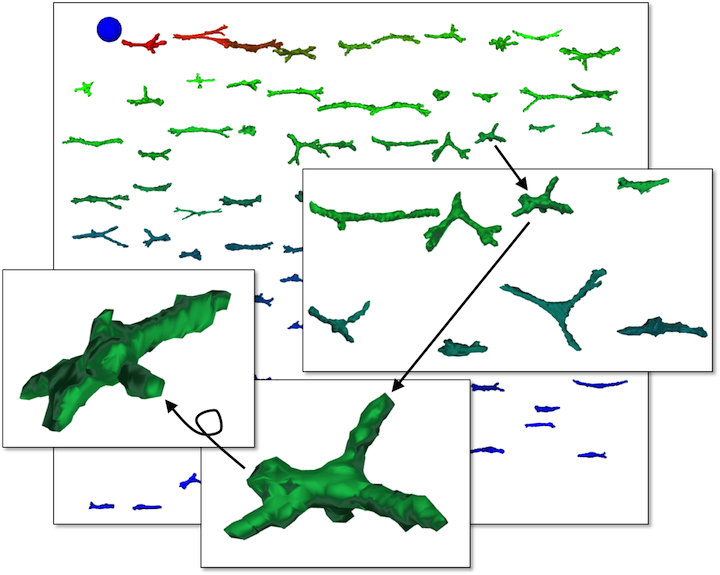
The 3D shapes obtained from actin or nucleus segmented images are displayed by selecting scaffolds of interest or individual cells on the top-left menu and clicking on the "Load Meshes" button. Different color-coding and sorting options are also available on the bottom-left menu.
A rapid cell shape comparison between several scaffolds can be easily performed since the tool is able to load all the meshes at the same time at a lower-resolution. More detailed analysis of a particular mesh is also possible by right clicking on a cell of interest which will load the 3D volume at a full resolution. Zoom-in and out are enabled through the mouse-wheel and the "+" and "-" keys. Further help is available by clicking on the "?" button next to the "Load Meshes" button.
Explanation of Color/Order Metrics:
- File Name: file name
- Volume: 3D cell volume
- Surface area: 3D cell surface area
- Z-Depth: 3D cell volumes were fit with a gyration tensor and the "square
roots of the principal moments of the gyration tensor" denoted the semi-
axis lengths [sqrt(L1), sqrt(L2), sqrt(L3)] of a characteristic
ellipsoid that was representative of each cell. Z-Depth is the caliper-
diameter parallel to the smallest axis [sqrt(L1)]. The caliper diameter,
also known as the Feret diameter, is the size of an object along a
specific direction.
- Sqrt(L1): 3D cell volumes were fit with a gyration tensor and the
“square roots of the principal moments of the gyration tensor” denoted
the semi-axis lengths of a characteristic ellipsoid that was
representative of each cell. Sqrt(L1) is the length of the shortest
semi-axis of the characteristic ellipsoid.
- Sqrt(L2): 3D cell volumes were fit with a gyration tensor and the
“square roots of the principal moments of the gyration tensor” denoted
the semi-axis lengths of a characteristic ellipsoid that was
representative of each cell. Sqrt(L2) is the length of the median-
length semi-axis of the characteristic ellipsoid.
- Sqrt(L3): 3D cell volumes were fit with a gyration tensor and the
“square roots of the principal moments of the gyration tensor” denoted
the semi-axis lengths of a characteristic ellipsoid that was
representative of each cell. Sqrt(L3) is the length of the longest
semi-axis of the characteristic ellipsoid.
The raw z-stacks, volumetric and mesh representations of segmentation results, and 3D shape features are accessible from the following link:
Peter Bajcsy
peter.bajcsy@nist.gov
Phone: 301.975.2958